Содержание
1. Что такое UNet++?
Мощная архитектура для сегментации медицинских изображений. Эта архитектура, по сути, представляет собой сеть кодировщик-декодер с глубоким обучением, в которой подсети кодера и декодера соединены серией вложенных слоев. Переработанные слои направлены на сокращение семантического разрыва между картами признаков подсетей кодировщика и декодера.

Давайте построим модель U-Net++ на основе UNet++: A Nested U-Net Architecture for Medical Image Segmentation
Нам нужно будет провести идентификацию клеточных ядер.
Почему ядра?
Идентификация клеточных ядер является отправной точкой для большинства анализов, потому что большинство из 30 триллионов клеток в организме человека содержат ядро, содержащее ДНК генетического кода, который программирует каждую клетку. Идентификация ядра позволяет исследователям идентифицировать каждую отдельную клетку в образце. А измеряя, как клетки реагируют на различные виды лечения, исследователь может понять основные биологические процессы.
2. Зависимости
Для сборки UNet++ нам понадобятся: библиотека Tensorflow (глубокое обучение), NumPy (численные вычисления) и skimage (обработка изображений). Также мы будем использовать цветовую палитру matplotlib.
import pandas as pd
import numpy as np
import zipfile
import os
import glob
import random
import sys
import skimage.io #Used for imshow function
import skimage.transform #Used for resize function
from skimage.morphology import label #Used for Run-Length-Encoding RLE to create final submission
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow.keras.layers import *
from tensorflow.keras.models import Model
from tensorflow.keras import backend as K
from tensorflow.keras.callbacks import LearningRateScheduler, ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from sklearn.model_selection import train_test_split
# Custom IoU metric
def mean_iou(y_true, y_pred):
prec = []
for t in np.arange(0.5, 1.0, 0.05):
y_pred_ = tf.to_int32(y_pred > t)
score, up_opt = tf.metrics.mean_iou(y_true, y_pred_, 2)
K.get_session().run(tf.local_variables_initializer())
with tf.control_dependencies([up_opt]):
score = tf.identity(score)
prec.append(score)
return K.mean(K.stack(prec), axis=0)
IMG_WIDTH = 256
IMG_HEIGHT = 256
IMG_CHANNELS = 3
print('Python :', sys.version.split('\n')[0])
print('Numpy :', np.__version__)
print('Skimage :', skimage.__version__)
print('Tensorflow :', tf.__version__)3. Параметры модели
Далее мы определим некоторые конфигурации для UNet++.
# learning rate
LR = 0.001
# Custom loss function
def dice_coef(y_true, y_pred):
smooth = 1.
y_true_f = K.flatten(y_true)
y_pred_f = K.flatten(y_pred)
intersection = K.sum(y_true_f * y_pred_f)
return (2. * intersection + smooth) / (K.sum(y_true_f) + K.sum(y_pred_f) + smooth)
def bce_dice_loss(y_true, y_pred):
return 0.5 * tf.keras.losses.binary_crossentropy(y_true, y_pred) - dice_coef(y_true, y_pred)
NUM_EPOCHS=254. Загрузка изображений.
Распаковываем данные. Загружаем и объединяем изображения из path/{id}/masks/{id}.png в пустой массив. Визуализируем.
# unpack the data
import zipfile
for name_data in ['test', 'train']:
tmp_zip = zipfile.ZipFile('../input/sf-dl-nucleus-detection/'+name_data+'.zip')
tmp_zip.extractall(name_data)
tmp_zip.close()
# to load the data, we will use the convenient skimage lib
def get_X_data(path, output_shape=(None, None)):
'''
Loads images from path/{id}/images/{id}.png into a numpy array
'''
img_paths = ['{0}/{1}/images/{1}.png'.format(path, id) for id in os.listdir(path)]
X_data = np.array([skimage.transform.resize(skimage.io.imread(path)[:,:,:3],
output_shape=output_shape,
mode='constant',
preserve_range=True) for path in img_paths], dtype=np.uint8) #take only 3 channels/bands
return X_data
def get_Y_data(path, output_shape=(None, None)):
'''
Loads and concatenates images from path/{id}/masks/{id}.png into a numpy array
'''
img_paths = [glob.glob('{0}/{1}/masks/*.png'.format(path, id)) for id in os.listdir(path)]
Y_data = []
for i, img_masks in enumerate(img_paths): #loop through each individual nuclei for an image and combine them together
masks = skimage.io.imread_collection(img_masks).concatenate() #masks.shape = (num_masks, img_height, img_width)
mask = np.max(masks, axis=0) #mask.shape = (img_height, img_width)
mask = skimage.transform.resize(mask, output_shape=output_shape+(1,), mode='constant', preserve_range=True) #need to add an extra dimension so mask.shape = (img_height, img_width, 1)
Y_data.append(mask)
Y_data = np.array(Y_data, dtype=np.bool_)
return Y_data
# Get training data
X_train = get_X_data('train', output_shape=(IMG_HEIGHT,IMG_WIDTH))
# Get training data labels
Y_train = get_Y_data('train', output_shape=(IMG_HEIGHT,IMG_WIDTH))
X_test = get_X_data('test', output_shape=(IMG_HEIGHT,IMG_WIDTH))
TRAIN_PATH = 'train/'
# Check training data
train_ids = next(os.walk(TRAIN_PATH))
f, axarr = plt.subplots(2,4)
f.set_size_inches(20,10)
ix = random.randint(0, len(train_ids[1]))
axarr[0,0].imshow(X_train[ix])
axarr[0,1].imshow(np.squeeze(Y_train[ix]))
axarr[0,2].imshow(X_train[ix])
axarr[0,3].imshow(np.squeeze(Y_train[ix]))
axarr[1,0].imshow(X_train[ix])
axarr[1,1].imshow(np.squeeze(Y_train[ix]))
axarr[1,2].imshow(X_train[ix])
axarr[1,3].imshow(np.squeeze(Y_train[ix]))
plt.show()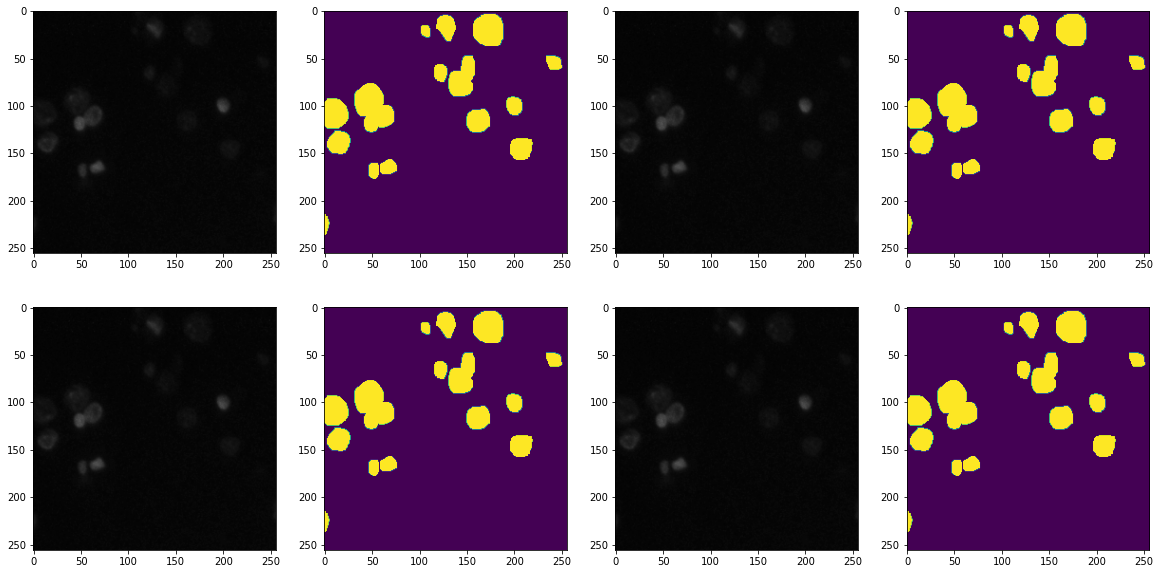
5. Аугментация изображений.
x_train, x_test, y_train, y_test = train_test_split(X_train, Y_train, test_size=0.1, random_state=13)
data_gen_args = dict(rotation_range=45.,
width_shift_range=0.1,
height_shift_range=0.1,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
vertical_flip=True,
fill_mode='reflect')
X_datagen = ImageDataGenerator(**data_gen_args)
Y_datagen = ImageDataGenerator(**data_gen_args)
test_datagen = ImageDataGenerator()
X_datagen_val = ImageDataGenerator()
Y_datagen_val = ImageDataGenerator()
X_datagen.fit(x_train, augment=True, seed=13)
Y_datagen.fit(y_train, augment=True, seed=13)
test_datagen.fit(X_test, augment=True, seed=13)
X_datagen_val.fit(x_test, augment=True, seed=13)
Y_datagen_val.fit(y_test, augment=True, seed=13)
X_train_augmented = X_datagen.flow(x_train, batch_size=15, shuffle=True, seed=13)
Y_train_augmented = Y_datagen.flow(y_train, batch_size=15, shuffle=True, seed=13)
test_augmented = test_datagen.flow(X_test, shuffle=False, seed=13)
X_train_augmented_val = X_datagen_val.flow(x_test, batch_size=15, shuffle=True, seed=13)
Y_train_augmented_val = Y_datagen_val.flow(y_test, batch_size=15, shuffle=True, seed=13)
train_generator = zip(X_train_augmented, Y_train_augmented)
val_generator = zip(X_train_augmented_val, Y_train_augmented_val)6. Построение модели
Построим архитектуру модели на Tensorflow
tf.keras.backend.clear_session()
nb_filter = [32,64,128,256,512]
# Build U-Net++ model
inputs = Input((IMG_HEIGHT, IMG_WIDTH, IMG_CHANNELS))
s = Lambda(lambda x: x / 255) (inputs)
c1 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (s)
c1 = Dropout(0.5) (c1)
c1 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (c1)
c1 = Dropout(0.5) (c1)
p1 = MaxPooling2D((2, 2), strides=(2, 2)) (c1)
c2 = Conv2D(64, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (p1)
c2 = Dropout(0.5) (c2)
c2 = Conv2D(64, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (c2)
c2 = Dropout(0.5) (c2)
p2 = MaxPooling2D((2, 2), strides=(2, 2)) (c2)
up1_2 = Conv2DTranspose(nb_filter[0], (2, 2), strides=(2, 2), name='up12', padding='same')(c2)
conv1_2 = concatenate([up1_2, c1], name='merge12', axis=3)
c3 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv1_2)
c3 = Dropout(0.5) (c3)
c3 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (c3)
c3 = Dropout(0.5) (c3)
conv3_1 = Conv2D(128, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (p2)
conv3_1 = Dropout(0.5) (conv3_1)
conv3_1 = Conv2D(128, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv3_1)
conv3_1 = Dropout(0.5) (conv3_1)
pool3 = MaxPooling2D((2, 2), strides=(2, 2), name='pool3')(conv3_1)
up2_2 = Conv2DTranspose(nb_filter[1], (2, 2), strides=(2, 2), name='up22', padding='same')(conv3_1)
conv2_2 = concatenate([up2_2, c2], name='merge22', axis=3) #x10
conv2_2 = Conv2D(64, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv2_2)
conv2_2 = Dropout(0.5) (conv2_2)
conv2_2 = Conv2D(64, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv2_2)
conv2_2 = Dropout(0.5) (conv2_2)
up1_3 = Conv2DTranspose(nb_filter[0], (2, 2), strides=(2, 2), name='up13', padding='same')(conv2_2)
conv1_3 = concatenate([up1_3, c1, c3], name='merge13', axis=3)
conv1_3 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv1_3)
conv1_3 = Dropout(0.5) (conv1_3)
conv1_3 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv1_3)
conv1_3 = Dropout(0.5) (conv1_3)
conv4_1 = Conv2D(256, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (pool3)
conv4_1 = Dropout(0.5) (conv4_1)
conv4_1 = Conv2D(256, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv4_1)
conv4_1 = Dropout(0.5) (conv4_1)
pool4 = MaxPooling2D((2, 2), strides=(2, 2), name='pool4')(conv4_1)
up3_2 = Conv2DTranspose(nb_filter[2], (2, 2), strides=(2, 2), name='up32', padding='same')(conv4_1)
conv3_2 = concatenate([up3_2, conv3_1], name='merge32', axis=3) #x20
conv3_2 = Conv2D(128, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv3_2)
conv3_2 = Dropout(0.5) (conv3_2)
conv3_2 = Conv2D(128, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv3_2)
conv3_2 = Dropout(0.5) (conv3_2)
up2_3 = Conv2DTranspose(nb_filter[1], (2, 2), strides=(2, 2), name='up23', padding='same')(conv3_2)
conv2_3 = concatenate([up2_3, c2, conv2_2], name='merge23', axis=3)
conv2_3 = Conv2D(64, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv2_3)
conv2_3 = Dropout(0.5) (conv2_3)
conv2_3 = Conv2D(64, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv2_3)
conv2_3 = Dropout(0.5) (conv2_3)
up1_4 = Conv2DTranspose(nb_filter[0], (2, 2), strides=(2, 2), name='up14', padding='same')(conv2_3)
conv1_4 = concatenate([up1_4, c1, c3, conv1_3], name='merge14', axis=3)
conv1_4 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv1_4)
conv1_4 = Dropout(0.5) (conv1_4)
conv1_4 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv1_4)
conv1_4 = Dropout(0.5) (conv1_4)
conv5_1 = Conv2D(512, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (pool4)
conv5_1 = Dropout(0.5) (conv5_1)
conv5_1 = Conv2D(512, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv5_1)
conv5_1 = Dropout(0.5) (conv5_1)
up4_2 = Conv2DTranspose(nb_filter[3], (2, 2), strides=(2, 2), name='up42', padding='same')(conv5_1)
conv4_2 = concatenate([up4_2, conv4_1], name='merge42', axis=3) #x30
conv4_2 = Conv2D(256, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv4_2)
conv4_2 = Dropout(0.5) (conv4_2)
conv4_2 = Conv2D(256, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv4_2)
conv4_2 = Dropout(0.5) (conv4_2)
up3_3 = Conv2DTranspose(nb_filter[2], (2, 2), strides=(2, 2), name='up33', padding='same')(conv4_2)
conv3_3 = concatenate([up3_3, conv3_1, conv3_2], name='merge33', axis=3)
conv3_3 = Conv2D(128, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv3_3)
conv3_3 = Dropout(0.5) (conv3_3)
conv3_3 = Conv2D(128, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv3_3)
conv3_3 = Dropout(0.5) (conv3_3)
up2_4 = Conv2DTranspose(nb_filter[1], (2, 2), strides=(2, 2), name='up24', padding='same')(conv3_3)
conv2_4 = concatenate([up2_4, c2, conv2_2, conv2_3], name='merge24', axis=3)
conv2_4 = Conv2D(64, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv2_4)
conv2_4 = Dropout(0.5) (conv2_4)
conv2_4 = Conv2D(64, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv2_4)
conv2_4 = Dropout(0.5) (conv2_4)
up1_5 = Conv2DTranspose(nb_filter[0], (2, 2), strides=(2, 2), name='up15', padding='same')(conv2_4)
conv1_5 = concatenate([up1_5, c1, c3, conv1_3, conv1_4], name='merge15', axis=3)
conv1_5 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv1_5)
conv1_5 = Dropout(0.5) (conv1_5)
conv1_5 = Conv2D(32, (3, 3), activation='elu', kernel_initializer='he_normal', padding='same') (conv1_5)
conv1_5 = Dropout(0.5) (conv1_5)
nestnet_output_4 = Conv2D(1, (1, 1), activation='sigmoid', kernel_initializer = 'he_normal', name='output_4', padding='same')(conv1_5)
model = Model([inputs], [nestnet_output_4])
model.compile(optimizer=tf.keras.optimizers.Adam(learning_rate=LR), loss=bce_dice_loss)
7. Запуск модели
Теперь мы можем запустить модель.
checkpoint = ModelCheckpoint('best_model.hdf5' ,
monitor = 'val_loss',
verbose = 1,
save_best_only=True,
mode = 'min',
save_weights_only=True,
save_freq='epoch'
)
reduce_lr = ReduceLROnPlateau(monitor='val_loss', factor=0.3,
patience=5, min_lr=0.00005)
callbacks_list = [checkpoint, reduce_lr]
# Fit model
history = model.fit(train_generator,
validation_data=val_generator,
steps_per_epoch=len(X_train)/(6),
validation_steps=10,
callbacks=callbacks_list,
epochs=NUM_EPOCHS,
verbose=1,)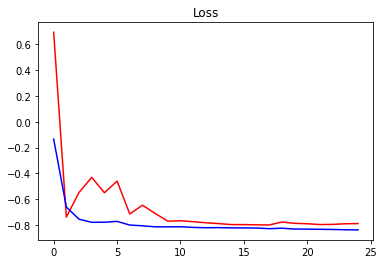
8. Визуализация результатов
Y_predict = model.predict(X_train, verbose=1)
train_ids = next(os.walk('train'))
test_ids = next(os.walk('test'))
# Check predict data
f, axarr = plt.subplots(2,3)
f.set_size_inches(20,10)
ix = random.randint(0, len(train_ids[1]))
axarr[0,0].imshow(X_train[ix])
axarr[0,0].set_title('Microscope')
axarr[0,1].imshow(np.squeeze(Y_predict[ix]))
axarr[0,1].set_title('"Predicted" Masks')
axarr[0,2].imshow(np.squeeze(Y_train[ix]))
axarr[0,2].set_title('"GroundTruth" Masks')
axarr[1,0].imshow(X_train[ix])
axarr[1,0].set_title('Microscope')
axarr[1,1].imshow(np.squeeze(Y_predict[ix]))
axarr[1,1].set_title('"Predicted" Masks')
axarr[1,2].imshow(np.squeeze(Y_train[ix]))
axarr[1,2].set_title('"GroundTruth" Masks')
plt.show()
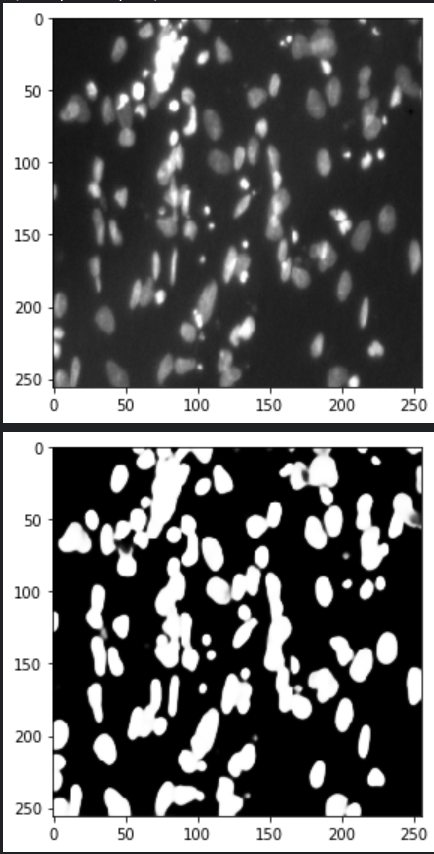
# Use model to predict test labels
Y_hat = model.predict(X_test, verbose=1)
Y_hat.shape
idx = random.randint(0, len(test_ids[1]))
print(X_test[idx].shape)
skimage.io.imshow(X_test[idx])
plt.show();
skimage.io.imshow(Y_hat[idx][:,:,0])
plt.show();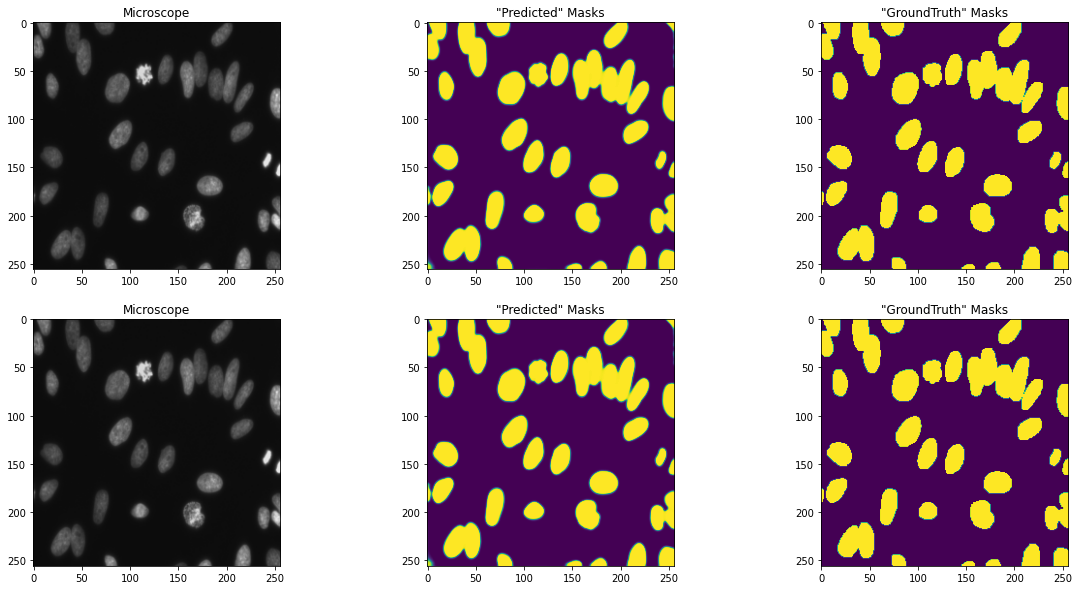
Смотрится неплохо.
Для удобства практического применения код доступен на Kaggle
enclis
UNet++ была предложена больше 4-х лет назад - ещё из тех времён, когда трансформеры ещё не заняли доминирующего положения во многих прикладных задачах. Конкретно для анализа медицинский изображений - обзор, ещё конкретней в области сегментации ядер клеток лучше всех пока себя показывает LViT.